The goal of the Small Worlds project is to develop a new multi-modal imaging capability for studying complex multi-agent processes in cells and systems of cells across physical and temporal scales. For example, understanding the detailed interactions amongst the synergistically functioning organisms, particularly bacteria and roots, will enable the development of models that will allow enhancing the growth and health of a wide range of plants. To create this new experimental capability, we will develop two major technological axes: multi-modal imaging in 3D, and multi-agent molecular sensor systems able to target several elements of a process at once. The combination of these, with supporting software for image reconstruction, volumetric data fusion, and quantitative analysis, will allow scientists to target complex processes in a wide range of biological systems. This contingent of capabilities will enable construction of dynamic experiments that are able to track and correlate interrelated molecular actors in complex processes, while providing detailed corroboration and supplementary data across physical scales with qualitatively different imaging modalities.
Approach: We propose to develop a 3D Snapshot Interferometric Holographic Microscope (3D-SIHM) capable of imaging whole live cells in fluorescence and/or brightfield modes simultaneously in 3D. This new type of microscope will be capable of obtaining the information required for reconstruction of 3D multi-scale volumetric information of complex systems in a single “snapshot” measurement. This dynamic 3D snapshot microscopy will be complemented by new correlative tomographic methods in two separate imaging modalities. Electron tomography will provide high resolution matching of dynamical processes to detailed cellular structure. X-ray tomography will enable larger-scale inter-cellular correlations to be developed so that cross-organismal processes may be studied. Supplementing these developments in three-dimensional multi-modal imaging will be the development of new multi-agent molecular systems technology.
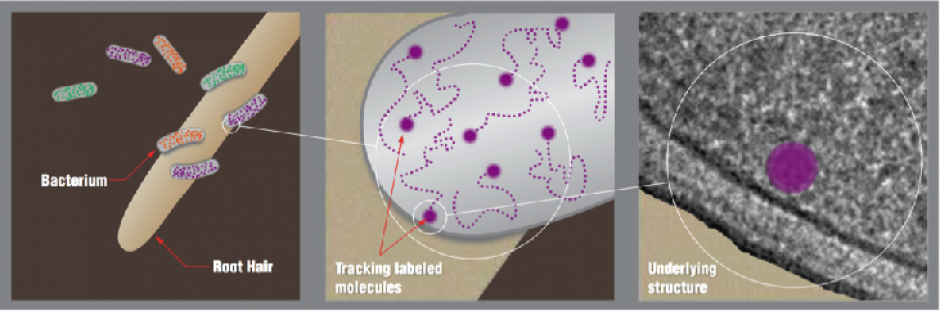
The image at the top of this page shows schematically how the Small Worlds workflow might be applied to a transport problem in the rhizosphere of the poplar tree. (Left) X-ray tomography will aid identification of target organisms in genetically marked sample. (Center) Dynamic 3D imaging will then study molecular interactions to uncover process mechanics. (Right) Electron tomography will correlate detailed dynamics with high-resolution cellular structure.
Impact: Small Worlds will create a platform for studying a range of complex processes in cellular and inter-cellular systems. This work will systematize creation of sensor systems capable of simultaneously tracking, sensing, and/or controlling several aspects of a complex process in a single experiment. It will also provide the means to correlate image volumes by providing markers that function across modalities.